The Human Brain Project studies the brain and its diseases. In the first approach, a realistic model of the brain is recreated based on detailed neuroscience and genetic data. The second is to collect large amounts of data from many aspects of a disease in one data format. The goal is to combine both approaches to link complex genetic and neurophysiological causes of psychiatric diseases with their associated complex phenotypes.
It has long been recognized that psychiatric diseases such as schizophrenia and depression cannot be explained monocausally, but that multifactorial explanations are required. At the level of research methodology, this means that isolated research approaches are not sufficient, but that methods are needed that can capture the combinatorics of multifactorial mechanisms (e.g., genetic, neurobiological, phenomenological level). The same applies to the study of the human brain with billions of neurons and neuronal connections.

The goal of the Human Brain Project is to be able to explore the complexity of the brain and its diseases. Two approaches are envisaged for this purpose: First, a digital model of the brain will be reconstructed based on neuroscientific and genetic data. Second, large amounts of data from many aspects of a disease should be brought into a uniform data format. The vision of this project is to combine these two approaches and to find new explanatory approaches by integrating genetic and neurophysiological features of psychiatric diseases and their associated complex phenotypes. So, in summary, it is an approach that deals with collecting and analyzing large amounts of data related to the human brain.
“Bottom-up approach and top-down approach
The bottom-up approach focuses on the goal of completely rebuilding the brain, i.e. digitizing it. On a methodological level, this is a data integration project comparable to the development of a digital atlas based on various data sources of different scales. Based on data from the “Blue Brain Project” [1], a digital model is being designed of the structure and functioning of the brain. In the top-down approach, data are collected regarding different characteristics of a disease (genetic and molecular level, behavioral level, etc.). These data are collected in clinics and partner universities, remain on the computers of the respective institutions and are analyzed by special software. Parameters of these analyses are then stored in a central neuroinformatics platform. From there, this data is passed on to other platforms:
- Simulation platform (=examination of healthy brain)
- Platform, which is about diseases

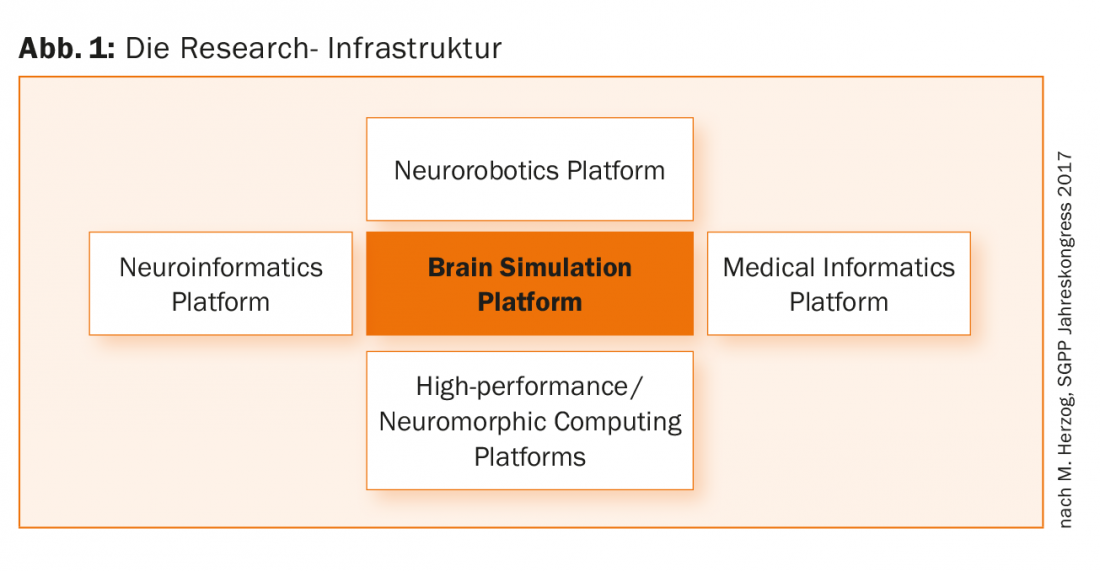
There are twelve subprojects within the Human Brain Project. For example, as part of Subproject 10 (“Neurorobotics”), a new web-based environment (“Neurobiotics Platform”) (Fig. 1) is available to scientists and other professionals that provides the ability to link brain models with detailed simulations of robots and their environments, and to use resulting neurorobotic systems for in silico experiments [2].
The medical informatics platform
The sub-project on research into mental illness (“The Medical Informatics Platform”) (Fig. 1) is about a better understanding of mental illness. It involves 14 partner institutions and 5 clinics, including a research group led by Ferah Kherif and Bogdan Draganski, both researchers and lecturers at the University of Lausanne.
Brain science and clinical diagnostics are to be linked and brought into a standardized format in terms of data technology. The goal is to better predict and explain complex diseases by integrating data on clinical (phenomenology), etiological, and biological features and matching these data with the digital model of the brain.
How is this implemented? One measures all available information about a particular disease at the genetic, molecular, or behavioral level. Machine learning algorithms are used to analyze this data. In this way, the network learns to find differences and form clusters within a huge data set.
For example, an analysis of a large dataset of Alzheimer’s disease patients [3] with data on genetic, molecular, and cognitive characteristics using this method resulted in the identification of distinct subgroups. This method can be used to study how certain mechanisms produce a certain phenomenology. In a next step, these findings could be used to develop subgroup-specific drugs. Since the Human Brain Project is a “Big Data” infrastructure with the goal of gaining access to a large amount of clinical data, ethical issues, such as the conditions of informed consent, must also be addressed [4].
Concrete application example: explanatory model for schizophrenia
Empirical studies show that there are abnormalities in various features at different levels in schizophrenic disorders and that genetic factors play a major role. For example, it was shown that the risk of developing schizophrenia at some point in life (“lifetime risk”) is 50% for a twin sibling of a person with the disease. Children of ill parents have a 13% risk of also developing schizophrenia. Empirical studies have found that schizophrenia patients are significantly slower on tasks with high demands on working memory compared to a control group of healthy individuals, and that neural activity patterns in the prefrontal cortex differ. In a test battery, several tests (e.g., Wisconsin Card Sorting Test, Visual Masking, Stroop, etc.) showed abnormalities. However, hardly any correlations could be found between results of the individual tests as well as “single nucleotide” polymorphisms and results in cognitive tests. This shows that it is a complex disease.
Herzog illustrated the combinatorial challenge of a multifactorial disease such as schizophrenia in simplified form using a thought experiment: assuming there are eight disease-relevant factors and each of these factors has two variants (e.g., “Up,” “Down”), there are 256 “Up”/”Down” constellations. Each individual component is unproblematic, the combination of the different variants is crucial. That all variants are “Up” or “Down” corresponds to a probability of less than 1%. This corresponds to the prevalence of schizophrenia in the general population. To study the mechanisms of complex diseases in more detail, large amounts of data are required, as well as analytical methods that can capture the combination of different features and form clusters. This is an example of one possible application of the Human Brain Project research methodology.
Planned research activities
In April 2018, there will be a call for proposals with more details on the conditions of participation in the Human Brain Project. One option is to conduct data collection in the clinical setting. It is envisaged that software for the analysis of data will be installed on the computers of the respective clinic and that only anonymized and encrypted metadata will be exported to computers outside. The original data therefore always remains on the clinic’s computers. The encrypted metadata should be made available to the public and accessible to interested persons. In the future, anyone who registers with the project will be able to find out, for example, how a certain protein concentration affects certain other parameters.
- Website The Human Brain Project
www.humanbrainproject.eu

Literature:
- The Blue Brain Project. A Swiss Brain Initiative. http://bluebrain.epfl.ch
- Falotico E, et al: Connecting Artificial Brains to Robots in a Comprehensive Simulation Framework: The Neurorobotics Platform. Front Neurorobot, 2017 Jan 25; https://doi.org/10.3389/fnbot.2017.00002
- Adaszewski S, et al: How early can we predict Alzheimer’s disease using computational anatomy? Neurobiol Aging 2013; 34(12): 2815-2826.
- Christen M, et al.: On the Compatibility of Big Data Driven Research and Informed Consent: The Example of the Human Brain Project. In: Mittelstadt B., Floridi L. (eds). The Ethics of Biomedical Big Data. Law, Governance and Technology Series 2016; 29. Springer, Cham.
Further reading:
- Amunts K, et al: The Human Brain Project: Creating a European Research Infrastructure to Decode the Human Brain: Neuron 2016; 92 (3): 574-581, https://doi.org/10.1016/j.neuron.2016.10.046
- Amunts K, et al. BigBrain: An Ultrahigh-Resolution 3D Human Brain Model. Science 2013; 340: 1472-1475, DOI: 10.1126/science.1235381, http://science.sciencemag.org/content/340/6139/1472
- Deco G, Kringelbach ML. Great Expectations: Using Whole-Brain Computational Connectomics for Understanding Neuropsychiatric Disorders. Neuron 2014; 84 (5): 892-905. https://doi.org/10.1016/j.neuron.2014.08.034
InFo NEUROLOGY & PSYCHIATRY 2017; 15(6): 46-48.