The use of the four multigene tests MammaPrint, Oncotype DX, EndoPredict and Prosigna may be useful in deciding on adjuvant chemotherapy. In which patients in particular should they be used?
Fortunately, in recent decades, mortality from breast carcinoma has declined. The reasons for this are the earlier diagnosis of breast carcinomas, new substances and also the frequent use of adjuvant chemotherapy. The question now arises whether therapy could be de-escalated in some patients [1].
Decision based on clinical and pathological risk factors
The decision as to which patient should receive adjuvant chemotherapy and which should not was previously based on clinical and pathological risk factors such as tumor size, lymph node status, tumor grade, proliferation rate, hormone receptor expression, and Her2 status.
In triplenegative (“basal-like”) carcinomas and in Her2-positive tumors, it is undisputed that adjuvant chemotherapy should be given. They usually have a high proliferation rate and tend to grow more aggressively. Both groups usually respond well to chemotherapy.
The decision is somewhat more difficult in well-differentiated estrogen receptor (ER)-positive luminal A and poorly differentiated ER-positive luminal B tumors, which are more difficult to distinguish. Thus, the proliferation rate, which was determined using a not always reliable KI-67 immunostaining, has been used for differentiation so far.
Multigene expression tests
Thus, in order to determine with greater certainty which patients should receive chemotherapy in addition to anti-hormonal therapy in the large group of hormone receptor-positive breast carcinomas, there is a need for better and more individualized risk prediction.
For example, the development of cDNA microarrays began about 20 years ago to study tumor gene expression in greater detail. With these arrays, the expression of about 500 genes could be determined simultaneously. It was found that the biologically different breast carcinoma types can also be distinguished from each other on the basis of the gene expression pattern.
Based on these studies, multigene expression tests have been developed for routine clinical use, which are limited to the most important genes necessary to distinguish prognostically different tumor types and allow individual therapy decisions.
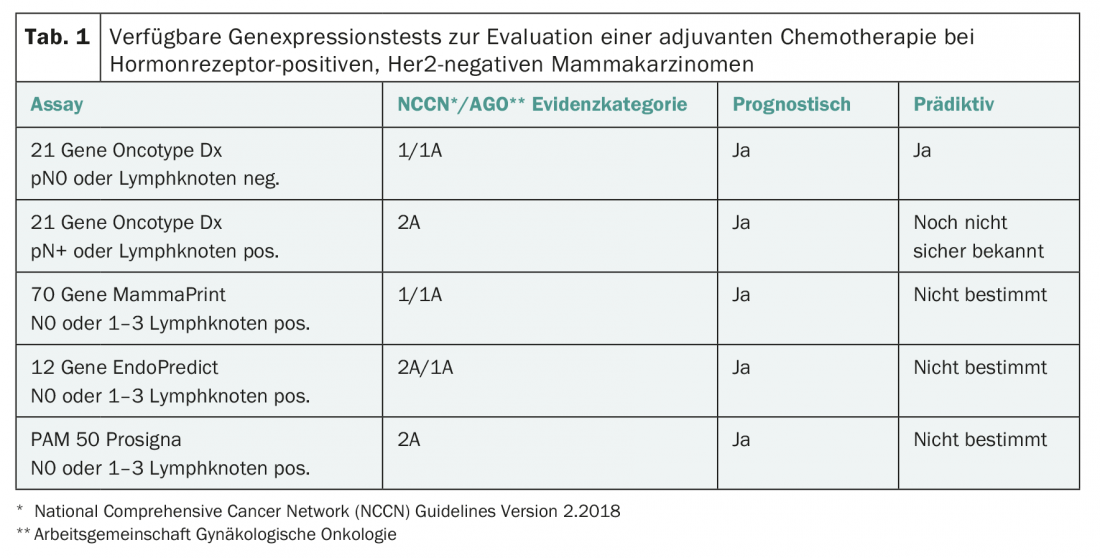
Four such multigene expression tests are now available in Switzerland (Tab. 1) [2–4]:
- MammaPrint: The test is performed centrally on frozen or formalin-fixed tumor material and examines the gene expression of 70 genes. MammaPrint distinguishes low from high prognostic risk.
- Oncotype DX: The test is performed centrally in the USA on paraffin-embedded material and examines the expression of 21 genes. It is prognostic for the risk of recurrence in the first five years and gives a numerical value between 0 and 100 (“recurrence score”) as a measure of the risk of recurrence. Based on Oncotype DX, conclusions about the efficacy of chemotherapies are also possible [2,3].
- EndoPredict: The test is performed decentrally, also in Switzerland, on formalin-fixed or paraffin-embedded tissue, tests the mRNA expression of 12 genes and includes clinicopathological parameters (tumor size, number of affected axillary lymph nodes). Thus, it is not a pure genetic test. The prognostic value for predicting early and late metastases has been validated.
- Prosigna: The test examines the expression of 50 genes and can also be performed locally with the appropriate laboratory equipment. It was developed to determine the biological subtype, but also has prognostic value by estimating the patient’s individual risk of relapse.
The prognostic significance of all four tests has been demonstrated in retrospective studies. Prospective data are now available for Oncotype DX and MammaPrint (TAILORx, Mindact [5,6]). All four tests are recommended as prognostic tests by international guidelines (ASCO, ESMO guidelines, AGO, St. Gallen Consensus [1]) and are now reimbursed by health insurance companies. However, it is not the case that the test results of the different tests correlate 100%. For example, a comparison of EndoPredict and Oncotype DX has been found to have a significant but rather moderate 76% agreement in results [7].
When is chemo useful?
The question now arises in which risk group defined by such a gene expression test adjuvant chemotherapy is appropriate. This question is being investigated in the currently published TAILORx and Mindact studies.
The TAILORx study [5] is a prospective study conducted using the 21-gene assay, Oncotype DX. Ten 273 women with hormone receptor-positive, Her2-negative breast carcinoma without lymph node involvement were included. The test result provides a “recurrence score” (RS) between 1 and 100, with a score between 1 and 11 predicting a low risk of recurrence where chemotherapy is not indicated, and a score >25 indicating a high risk where chemotherapy is indicated. Ambiguity prevailed in women with a median risk between 11 and 25, affecting the majority of patients (69%). This group of 9719 women was randomized into two study arms with and without chemotherapy. All patients received adjuvant anti-hormonal therapy. The observation period averaged nine years. After this period, both treatment groups had similar rates of disease-free survival, 83.3% in the group treated exclusively with anti-hormones and 84.3% in the group that also received chemotherapy. Regarding overall survival, there was no difference (93.9% vs. 93.8%), especially in women older than 50 years. Thus, in patients older than 50 years and with an RS of 0-25, chemotherapy is not expected to provide additional benefit. Among younger patients, particularly those with RS of 21-25, approximately 6.5% benefited from chemotherapy.
The Mindact study [6] was performed with the 70-gene signature MammaPrint. This test distinguishes tumors with low genetic risk from tumors with high genetic risk for the occurrence of distant metastases after five and ten years. Mindact is a phase III trial evaluating the utility of a microarray (MammaPrint) as additional information to clinical risk factors in making individualized decisions about the use of adjuvant chemotherapy. In 6693 women, MammaPrint was used to determine genetic risk and “Adjuvant Online” was used to determine clinical risk, including traditional risk factors. When clinical and genetic risk matched, treatment was clear: patients with low genetic and low clinical risk received no chemotherapy, and patients with high genetic and high clinical risk received chemotherapy. Patients at discordant risk, i.e., high genetic and low clinical or vice versa, were randomized into two treatment groups with and without chemotherapy in addition to antihormonal therapy. It was found that the administration of chemotherapy did not make a significant difference in the discordant groups with respect to disease-free survival. Looking at the group of 1550 patients with high clinical and low genetic risk, there is only a 1.5% difference in distant recurrence free survival (DRFS) after five years in favor of chemotherapy. Patients with low clinical and high genetic risk had no benefit from chemotherapy with respect to DRFS. Thus, one can conclude from this study that only patients with high clinical risk should be offered genetic testing and then patients with low genetic risk can be spared chemotherapy unless the patients insist on chemotherapy to minimize the risk of recurrence by another 1.5%. However, these patients then also do not need a genetic test.
St. Gallen Consensus Meeting 2017
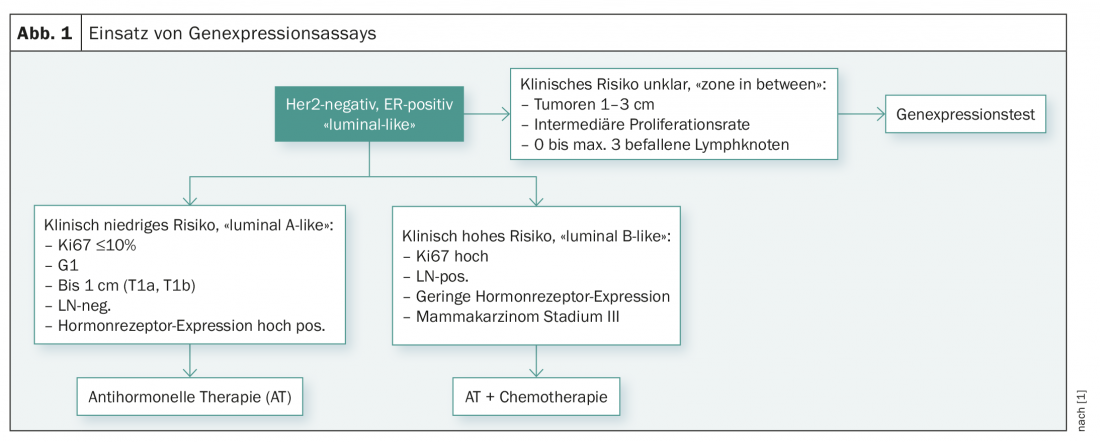
At the 2017 St. Gallen Consensus Meeting in Vienna, these studies and the use of multigene testing were also discussed [1]. The experts currently saw the indication of genetic testing only in answering the question whether a patient needs adjuvant chemotherapy or not. According to the current consensus recommendation, genetic testing should not be performed in cases of low clinical risk (pT1a/b, G1, ER high, N0, low proliferation rate) and clearly high clinical risk (low hormone receptor expression, G3, high proliferation rate, involvement of many lymph nodes or stage III). The expert panel considered genetic testing to be useful only in patients with a clinical risk profile that was not entirely clear (tumor size 1-3 cm, involvement of 0-3 axillary lymph nodes, and intermediate proliferation rate) (Fig. 1).
Literature:
- Curigliano G, et al: De-escalating and escalating treatments for early-stage breast cancer: the St. Gallen International Expert Consensus Conference on the Primary Therapy of Early Breast Cancer 2017. Annals of Oncology 2017; 28: 1700-1712.
- Paik S, et al: A Multigene Assay to Predict Recurrence of Tamoxifen-Treated, Node-Negative Breast Cancer. N Engl J Med 2004; 351(27): 2817-2826.
- Paik S, et al: Gene Expression and Benefit of Chemotherapy in Women With Node-Negative, Estrogen Receptor-Positive Breast Cancer. J Clin Oncol 2006; 24: 3726-3734.
- Markopoulos C, et al: Clinical evidence supporting genomic tests in early breast cancer: Do all genomic tests provide the same information? EJSO 2017; 43: 909-920.
- Sparano JA, et al: Adjuvant chemotherapy guided by a 21-gene expression assay in breast cancer. N Engl J Med 2018; 379: 111-121.
- Cardoso F, et al: 70-Gene Signature as an Aid to Treatment Decisions in Early-Stage Breast Cancer. N Engl J Med 2016; 375(8): 717-729.
- Varga Z, et al: Comparison of EndoPredict and Oncotype DX Test Results in Hormone Receptor Positive Invasive Breast Cancer. PLOS ONE 2013; 8(3): e58483.
- Lux MP, et al: Budget Impact analysis of gene expression tests to aid Therapy decisions for breast cancer patients in Germany. The Breast 2018; 37: 89-98.
Further reading:
- Sørlie T, et al: Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. PNAS 2001; 98(19): 10869-10874.
InFo ONCOLOGY & HEMATOLOGY 2018; 6(6): 26-29.