A summary of the current situation of pharmacogenetics in psychiatry – with practical advice for its application, as a revision of the analysis list has come into force since the beginning of 2017. Under certain conditions, this allows for reimbursement of pharmacogenetic testing.
Pharmacogenetics in psychiatry may soon celebrate its 50th birthday. Scandinavian authors had already found in 1969 that after treatment with nortriptyline, plasma levels of this noradrenergic tricyclic antidepressant were very similar within monozygotic but not dizygotic twin pairs, leading to the conclusion that the variability in plasma levels had a genetic background [1]. At the same time, this work also already indicated the occurrence of pharmacokinetic interactions that must be called environmental factors, in that the similarity of nortriptylink concentrations within pairs was not observed in the monozygotic twins treated with additional drugs.

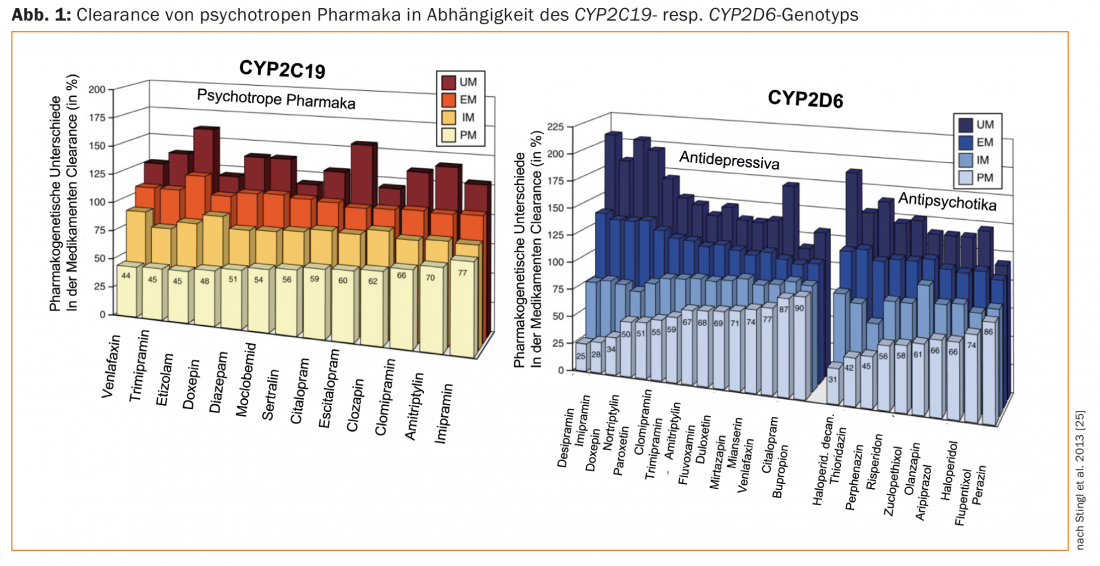
In 1977, the next important step was the description of different phenotypes, namely “efficient” (EM) or “normal” metabolizers and “non-metabolizers” (n.m., now called poor metabolizers, PM) of the antihypertensive drug debrisoquine [2]. A genetic deficiency of debrisoquine hydroxylase, which is now known as the genetically polymorphic enzyme cytochrome P-450 2D6, or CYP2D6, was suspected in PM based on pharmacokinetic data. By means of modern DNA analysis techniques, the presentation of genotypes was then achieved in 1988, namely the first molecular biological evidence of a genetic polymorphism, using CYP2D6 as an example [3]. In 1981, exceptionally high plasma levels of nortriptyline were measured in a nortriptyline-treated patient who was PM on the pharmacogenetic debrisoquin test. In addition, the patient suffered severe side effects despite usual dose (75 mg/day). Lowering the dose to 25 mg/day produced clinical improvement and resolution of side effects [4]. This historical case study is considered the first psychopharmacogenetic, clinically relevant description. Since then, numerous studies have supported the hypothesis that, in addition to environmental factors and factors tied to the individual patient, genetic factors are also responsible for interindividual differences in the pharmacokinetics and pharmacodynamics of psychotropic drugs (Fig. 1) [5]. In particular, numerous genetic variants have been identified in the enzymes involved in the metabolism of these drugs and their influence on enzyme activity has been characterized [6,7]. Thanks to the collaboration between academia and industry in the field of pharmacogenomics-pharmacogenetics, there are already genetic biomarkers suitable for implementation of personalized therapy in practice [7,8].
This paper summarizes the current situation of pharmacogenetics in psychiatry. It contains practical advice for their application, as recently pharmacogenomics in Switzerland has undergone an important innovation: On January 1, 2017, a revision of the list of analyses came into force, allowing reimbursement of pharmacogenetic tests by the basic insurance under certain conditions.
General importance of pharmacogenomics pharmacogenetics.
Under the name pharmacogenomics, a discipline has developed that studies the effect of interindividual genetic differences on pharmacokinetics, therapeutic effects, as well as side effects of drugs [9]. Pharmacogenomics and pharmacogenetics are often used interchangeably. Pharmacogenetic biomarkers allow to estimate the therapeutic response to a drug or its toxicity in an individual patient. These are often genetic variants of enzymes of drug metabolism or of transport proteins, of receptors responsible for drug action, but also of the major histocompatibility complex (human leukocyte antigens, HLA; see the following example carbamazepine).
In clinical practice, the following roles are attributed to pharmacogenetic testing [10]:
- Avoiding overdoses that lead to adverse drug reactions
- Avoiding underdoses that lead to insufficient therapeutic effect.
- Avoiding use of medications in hypersensitive, risk-exposed patients.
- Improve differential diagnosis, e.g., to establish causality of an adverse drug reaction.

The overall goal is to tailor drug therapies to an individual patient’s genetic profile using insights gained from pharmacogenomics pharmacogenetics. In this regard, other authors refer to “stratified pharmacotherapy” [11]. In this context, in order to contribute significantly to their implementation, but also to promote research, the Swiss group “Pharmacogenomics and Personalized Therapy” was recently founded as a section of the Swiss Society for Clinical Pharmacology and Toxicology (SGKPT) (Box “Swiss Group of Pharmacogenomics and Personalised Therapy”). It must be specified that pharmacogenetics does not usually study genes responsible for the development of diseases [12]. Pharmacogenomics studies gene variants that affect the areas of pharmacokinetics and pharmacodynamics of a drug. Drug targets are, for example, receptors or neurotransmitter transport proteins. Depending on the genetic variant present, drugs interact with them differently in pharmacological terms. Thus, drugs may differ interindividually in their pharmacodynamics due to differences in the genetic profiles of treated patients. In this regard, there are numerous studies on the importance of genetic variants of neurotransmitter transporter proteins (e.g., 5-HT transporter) and receptor proteins (e.g., dopamine receptor) for the clinical effects of psychotropic drugs [13,14]. However, genotyping in this area is currently not recommended in daily practice due to insufficient evidence [15].
The situation is different in the area of pharmacokinetics-pharmacogenomics, where a tentative implementation is already taking place. Many authors and committees recommend genotyping of drug-metabolizing enzymes and, more recently, drug transport proteins in clinical practice. The four stages in the fate of a drug in the organism are absorption, distribution, metabolism and elimination (ADME). In the past, it was assumed that drugs spread in the organism by means of simple diffusion. In the meantime, however, transport proteins have been characterized that accelerate the passage of active substances through membrane and cell barriers, e.g. from the intestine into the blood and through the blood-brain barrier. Uptake into the liver, where much of the drug metabolism takes place, is also mediated by transport proteins. One of these is the P-glycoprotein (PgP), which, among other things, acts as an efflux transporter to ensure that drugs and other xenobiotics, insofar as they are substrates of PgP, are rapidly removed from the CNS and thus cannot reach relevant intracerebral concentrations. The ABCB1 gene, which codes for the PgP protein, has numerous genetic variants. As a result of this genetic polymorphism, there are individuals in whom the function of PgP as a transport protein for certain substrates is exercised only to a limited extent or not at all, thus affecting their pharmacokinetics and clinical effect [16].
As mentioned at the beginning, the cytochrome P-450 enzyme system also plays an important role in the metabolism of pharmaceuticals. Several forms such as CYP2B6, CYP2C9, CYP2C19, and CYP2D6 exhibit genetic polymorphism. A rough distinction is made between PM with no active gene copy, EM with two active gene copies – they are now referred to as “normal metabolizers” [17] – intermediate metabolizers (IM) with, for example, only one active gene copy, and ultrafast metabolizers (UM) with more than two active gene copies as a consequence of gene multiplication. Depending on the genetically determined metabolic status, extremely high (in PM), “normal” (in EM, IM) or extremely low (in UM) drug plasma levels are measured. There is therefore an increased risk of adverse drug reactions (in PM) or nonresponse (in UM) to medication at usual doses [18]. For drugs that are converted from an inactive form to the active agent by an enzyme with a genetic polymorphism, the risk groups are inverted (nonresponse in PM; risk of side effects in UM). For example, the CYP2D6 substrate codeine is a precursor (a “prodrug”) of morphine: in UM, the risk of morphine intoxication is increased after code administration [19].
Information sources for recommendations and guidelines
The treating physician bases his treatment strategy, among other things, on the information accessible in the “drug information” (SPC: Summary of product characteristics). This contains information on the metabolism, pharmacokinetics and mechanisms of action of the drugs. Several drugs now include evidence of the influence of pharmacogenetic variants. Despite recommendations in the scientific literature, it is extremely rare for drug information to explicitly recommend pharmacogenetic testing. The drug/gene pairs carbamazepine/HLA-A*31:01 and carbamazepine/HLA-B*15:02 are an exception (in the pharmacodynamic spectrum). Carriers of the HLA-A*31:01 or HLA-B*15:02 alleles have a significantly increased risk of severe dermatologic adverse events compared with noncarriers. Therefore, in patients in whom carbamazepine therapy is planned, genotyping should be performed to reduce, among other things, the risk for the occurrence of Stevens-Johnson syndrome. However, the frequency of these risk variants varies in different ethnic groups, which is why, for example, only HLA-A*31:01 genotyping is recommended for individuals of European origin, as described in detail under “Carbamazepine” [20]. In contrast, the HLA-B*15:02 variant is common only in certain Asian populations, and genotyping is recommended only in patients of Asian origin.
Another exception concerns a pharmacokinetic-pharmacogenetic genotyping, namely the CYP2D6/ pair.Atomoxetine, a drug prescribed for the treatment of ADHD (“Attention deficit hyperactivity disorder”), which is a substrate of the enzyme. The “Drug Information” specifies: “About 7% of all Caucasians have a genotype corresponding to a defective CYP2D6 enzyme (so-called CYP2D6 “poor metaboliser”). Patients with this genotype have many times increased exposure to atomoxetine compared to patients with a functioning enzyme. “Poor metabolisers” therefore have an increased risk of side effects. For patients with a known “poor metaboliser” genotype, a lower starting dose and slower titration should be considered.” Despite these recommendations for action, the obvious genotyping before starting therapy is not explicitly recommended in the drug information. For the same drug, on the other hand, pharmacokinetic data allow the assumption that in UM (CYP2D6) patients the risk of a lack of effect (non-response) to atomoxetine is high [21].
For other drugs, such as aripiprazole, although the drug information mentions that this antipsychotic is metabolized by CYP2D6 and therefore its biotransformation is greatly reduced in PM, there is no information or recommended action on the potential benefit of genotyping. However, several author groups and organizations have now published guidelines with recommendations to assist the treating physician in interpreting pharmacogenetic test results and in making decisions about further treatment involving pharmacogenetic information.
Very helpful in this regard are organizations such as the Clinical Pharmacogenetics Implementation Consortium (CPIC) and the PharmGKB hosted by Stanford University. (The Pharmacogenomics Knowledgebase). They collect and analyze the scientific evidence in the field, publish recommendations for drug selection and drug dose adjustment based on pharmacogenetic test results and information on the relationship between specific genes and gene variants and the clinical effect on drug treatment outcome. PharmGKB also provides online interpretation tools where, after entering a patient’s genotype, recommendations relevant to that genotype are displayed directly.
This shows that it is not simply necessary to determine for a group of drugs with similar chemical structures or the same indication, but for each individual drug on the basis of experimental data and clinical studies, how relevant certain genetic variants are and whether genotyping is useful at all. Therefore, drug/gene pairs must be defined for which genotyping according to specific criteria will provide treatment-relevant, clinically directional information.
Evidence levels
PharmGKB has defined different levels of evidence that reflect the strength of scientific data supporting an association between genetic variants and a particular treatment outcome (effect, side effects) for individual gene/drug pairs (see box).

Pharmacogenetic studies are difficult to implement because, depending on the frequency of the alleles studied in a population or the side effect studied, a very large number of patients must be included in the study to obtain statistically relevant results. PharmGKB has therefore defined four different levels of evidence [22,23]. Each clinical annotation (“annotation”) thus contains information on the level achieved for the drug/gene pair in question.
Table 1 shows a compilation of the drugs most relevant to psychiatry, their proteins involved in metabolism and transport, and the clinical evidence levels of the associations. Ideally, the levels of evidence should also be reflected in the recommendations in the drug information. In addition, the U.S. Food and Drug Administration (FDA) has published a list of 200 drugs whose drug information provides information on pharmacogenomic biomarkers.
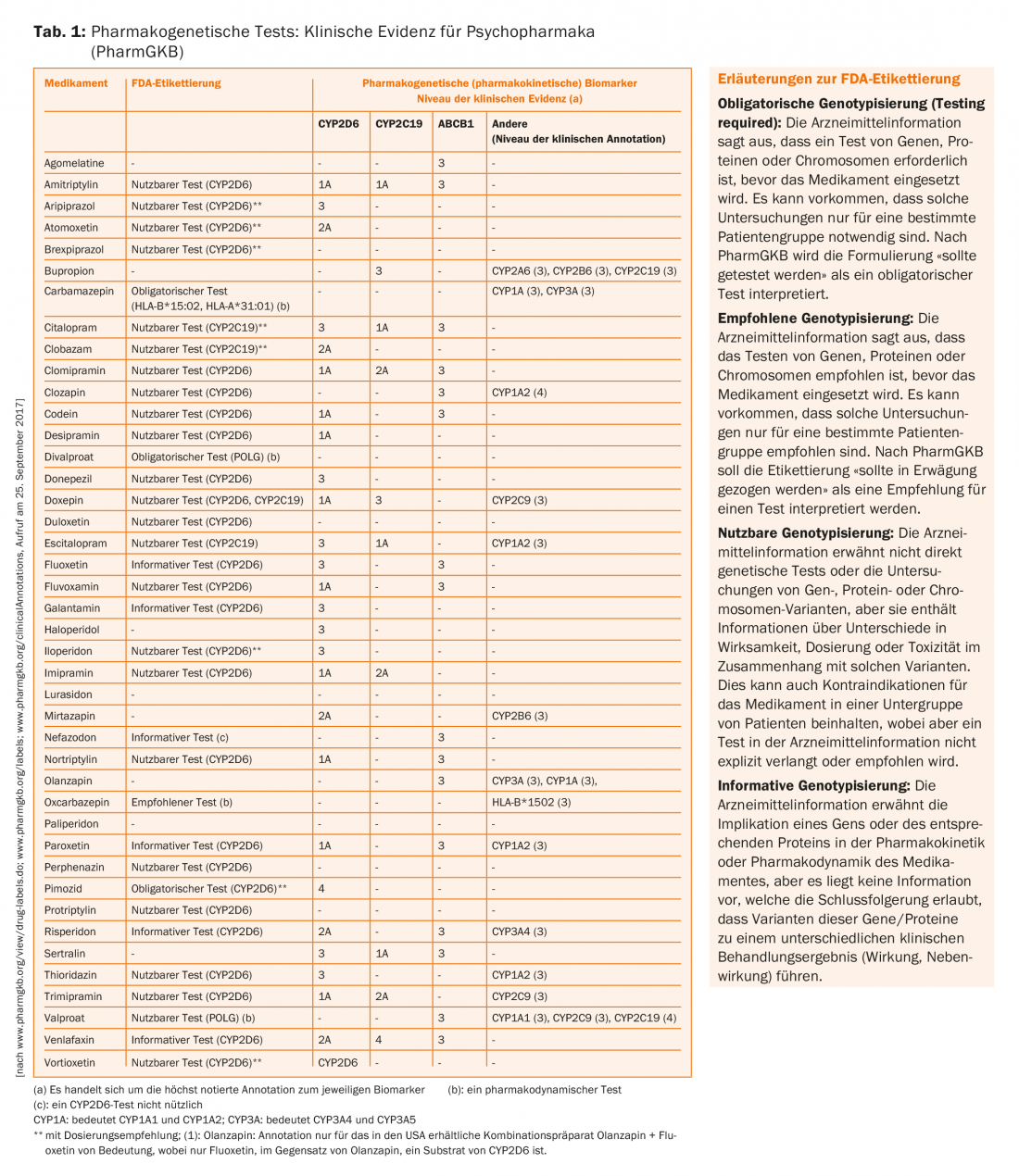
With 26 biomarkers, psychiatry ranks second only to oncology in terms of frequency (Table 1) [24]. It must be emphasized that in oncology, very often the genetics of the tumor is studied to estimate the effect of a drug (e.g., certain mutations in the EGF receptor must be present in tumor cells for gefitinib to work), but this usually differs from the genetics of the patient. In contrast, pharmacogenetic questions in the metabolism and transport as well as in the action of non-oncological drugs use the genetic information of the patient’s somatic cells.
Table 1 also presents annotations of FDA drug information for psychotropic drugs as summarized by PharmGKB. Corresponding annotations, but less complete and not presented here, also exist from the European Medical Agency (EMA), the Japanese Pharmaceutical and Medical Devices Agency (PMDA), Health Canada Santé Canada (HCSC), and other agencies. The following can be summarized from this compilation:
- For drug/gene pairs such as carbamazepine/HLA-A*31:01 allele and carbamazepine/HLA-B*15:02 allele, but also for certain antidepressants (e.g., amitriptyline)/CYP2D6 and/or CYP2C19, there are recommendations at the highest level (1A)
- Drugs/gene pairs related to transporter proteins such as ABCB1 (PgP) (e.g., citalopram/ABCB1) only reach evidence level 3.
- For virtually all drug/gene pairs, pharmacogenetic testing is indicated as “actionable,” i.e., usable or directional, but not necessarily mandated or classified as mandatory. Only for carbamazepine, divalproate (but strangely not for valproate – it could be the consequence of a lack of data from clinical trials to reach this recommendation level) and the antipsychotic pimozide, which is no longer approved in Switzerland, is a pharmacogenetic test designated as “required” by the FDA.
Recommendations for pharmacogenetic testing in psychiatry.
Reviews on the pharmacogenetics of psychotropic drugs and guidelines for pharmacogenetic testing in psychiatry deal in majority with CYP2D6 and CYP2C19 genotyping ofpatients treated with antidepressants and antipsychotics [25–29]. The drugs differ in their properties as substrates of these enzymes. This explains the significant differences in dose recommendations between different drugs considering the genotypes of the patients (UM, EM, IM, PM; Fig. 1; [25]). For example, for the near-exclusive CYP2D substrate paroxetine, the dose should be reduced to only about 50% of the usual dose for a PM (CYP2D6), and increased by over 50% for a UM. In contrast, this enzyme plays a minor role in citalopram, so dose adjustment is not necessary depending on the CYP2D6 genotype. In contrast, CYP2C19 plays a major role in the metabolism of this antidepressant – depending on the CYP2C19 genotype, the dose should be increased (UM) or decreased (PM) (Fig. 1).
Special attention deserves the recommendation of renowned Swiss authors who, on behalf of the Swiss Society for Anxiety and Depression (SGAD), the Swiss Society for Biological Psychiatry (SGBP) and the Swiss Society for Psychiatry and Psychotherapy (SGPP), have recommended genotyping of ABCB1 Recommend in all depressed patients who are prescribed therapy with antidepressants but experience insufficient therapeutic effect. [30]. This recommendation has public health and economic significance, as it may result in thousands of depressed patients being genotyped annually, whether or not they are treated with a PgP substrate.
Some antidepressants are substrates of PgP (citalopram), but others are not (mirtazapine). In depressed patients treated with antidepressants, a retrospective study found that genetic polymorphism of the ABCB1 genewas associated with treatment response in those treated with the PgP substrates amitriptyline, paroxetine, venlafaxine, or citalopram [31]. Of particular importance here was the distribution of genotypes of the ABCB1 variant(single nucleotide polymorphism, SNP) rs2032583. There were significantly fewer unremitted patients among carriers of the C allele of this variant (25%) than among patients who were not carriers of the C allele (62%). Similar results were observed in calculations related to the SNP rs2235015. Among carriers of the T allele of this variant, there were significantly fewer nonremitters compared to noncarriers. Such associations were not observed in a comparison group of patients not treated with PgP substrates.
As a limitation, it should be mentioned that a single antidepressant was prescribed for this group, namely mirtazapine. In a meta-analysis, the same team concluded that SNPs rs2032583 and rs2235015 were significantly associated with clinical effect [32], while analysis of other SNPs showed no such association. At the same time, however, researchers in another study concluded that maintaining therapeutic plasma levels of antidepressants is a necessary measure in addition to ABCB1 genotyping [33].
Some authors of the above meta-analysis recently published another, much more complete analysis of 32 studies on the association of the ABCB1 polymorphismand the clinical effect (therapeutic effect, tolerance) of antidepressants [32]: The highest associations are obtained when rs2032583 and rs2235040 are analyzed, but they are already weaker when rs2032582 is considered, and even weaker when other SNPs such as the above-mentioned rs2235015 are considered. Overall, the authors of the analysis concluded that the studies to date are insufficient to prove a clinical benefit of ABCB1 genotypingin the drug therapy of depression [34].
Further clinical studies are called for, as well as those demonstrating functional significance of these ABCB1 variantslocated outside the protein-coding region of the gene. This could be investigated with imaging techniques to find out whether patients with different ABCB1 genotypes or variant combinations (haplotypes) experience different uptake of the drugs in the brain after administration of antidepressants. An independent analysis of the literature confirms that, at this time, the general introduction of ABCB1 genotypinginto clinical practice is premature [16]. This conclusion was also recently reached by members of the Drug Commission of the German Medical Association [35].
For the practicing physician, this dispute is confusing, especially since one group of authors doubts that amitriptyline is even a Pgp substrate in humans based on their research findings [36]. It is also not readily apparent which gene variants are being investigated by the specialist laboratories offering pharmacogenetic analyses of ABCB1 . Routine ABCB1 genotyping of a majority of drug-treated depressed patients is therefore clearly not recommended at present, especially since studies on the sensitivity and specificity of the test are also lacking.
Significance of the January 1, 2017 regulation for pharmacogenetic testing.
On 1. On January 1, 2017, a revised ordinance came into force in Switzerland, according to which pharmacogenetic tests are covered by the compulsory health insurance if certain conditions are met (ordinance of the EDI of 29. September 1995 on benefits in compulsory health care insurance (KLV, as of 1. January 2017). Of practical importance is that these analyses are now included in the Federal List of Analyses (Annex 3 of the KLV). Pharmacogenetic testing is addressed in the July 1, 2017 analysis list under items 2150.10, 2250.10, 2271.01, 2547.01 (reviewed 7/14/2017). A distinction must be made between two conditions:
- A small number of pharmacogenetic tests can be prescribed by all physicians, regardless of their specialty title, and reimbursed by health insurers. A list of these analyses in connection with specified drugs (i.e., they are drug/gene pairs in each case), is provided by the Swiss Society for Clinical Pharmacology and Toxicology (SGKPT, compiled 9.6.2016, version 1):
- Abacavir (HLA-B*57:01);
- Carbamazepine (HLA-A*31:01 and HLA-B*15:02);
- 6-mercaptopurine, azathioprine (TPMT);
- 5-fluorouracil, capecitabine (DPYD);
- Irinotecan (UGT1A1).
This list is updated annually by the SGKPT, which bases its choice of gene/drug pairs on both the current scientific literature and evidence obtained through clinical experience. Of importance to psychiatry and neurology is that any psychiatrist, neurologist, or even primary care physician can (and should) perform genotyping of HLA-A*31:01 and HLA-B*15:02 (in patients of Asian descent) when a patient is newly scheduled for carbamazepine treatment, and health insurers will reimburse the patient for the cost of the test.
2. for pharmacogenetic tests to be prescribed for drug/gene pairs other than those indicated above, the following two limitations in the analysis list are particularly important:
- Limitation 1: “Only when there is an indication for the administration of a drug or when a drug-related side effect or reduced or absent therapeutic efficacy occurs during treatment with a drug for which there is a scientifically proven association between significant drug-related side effects (including toxic effects) or reduced or absent therapeutic efficacy and the gene mutations studied.”
- Limitation 4: “For drugs that are not on the list of the SGKPT, prescription of the analysis only by physicians with a federal continuing education title in clinical pharmacology and toxicology according to the Federal Law of June 23, 2006 on the University Medical Professions (Medical Professions Act, MedBG; SR 811.11)”.
Also noteworthy from an ethical perspective is Limitation 3, which states that the tests must not be used to make a diagnosis or to look for a predisposition to a genetic disease.
Practical hints
This paper shows that pharmacogenomics will have increasing importance in psychiatry for the planning and delivery of pharmacotherapy. As a result of the revision of the analysis list, pharmacogenetic tests relevant in psychiatry are available at the expense of mandatory health insurance if a specialist in clinical pharmacology and toxicology has determined the need for the test. Appropriate knowledge is necessary to plan pharmacogenetic tests and to interpret their results optimally. This topic should therefore be given increased attention in residency training in psychiatry and psychotherapy, and in continuing education and training programs [37–39]. Educational sessions for the general public are also necessary to promote general understanding of pharmacogenomics and to best achieve its implementation [40]. Recommended is literature that not only summarizes the genetic principles, but also includes guidance for practical procedures and clinical examples [11]. Moreover, in view of the difficult subject matter and as a consequence of the new regulation, collaboration with clinical pharmacologists and toxicologists is particularly valuable. Also, collaboration with laboratory professionals trained in genetics is recommended for practical execution.
Indications for pharmacogenetic testing include: Higher or lower plasma levels of drugs than those normally expected at the administered dose, suggesting that the patient has a genetic peculiarity in his metabolism. The drug must therefore be a substrate of the genetically polymorphic cytochrome P-450 enzyme to be tested. When a patient is comedicated with inhibitors, it should also be considered that as a consequence of drug interaction, high plasma levels may mimic a genetic deficiency of metabolism. Similarly, comedication with an enzyme-inducing drug may result in a decreased plasma level, which is not genetic. Pharmacogenetic testing should therefore generally be considered as an adjunct to therapeutic drug monitoring (TDM) (Fig. 2). This approach is also recommended by the Arbeitsgemeinschaft für Neuropsychopharmakologie und Pharmakopsychiatrie (AGNP), which is currently reissuing a “Consensus Guideline” for TDM: it describes the optimal use of TDM alone, but also in combination with pharmacogenetic tests [41,42].
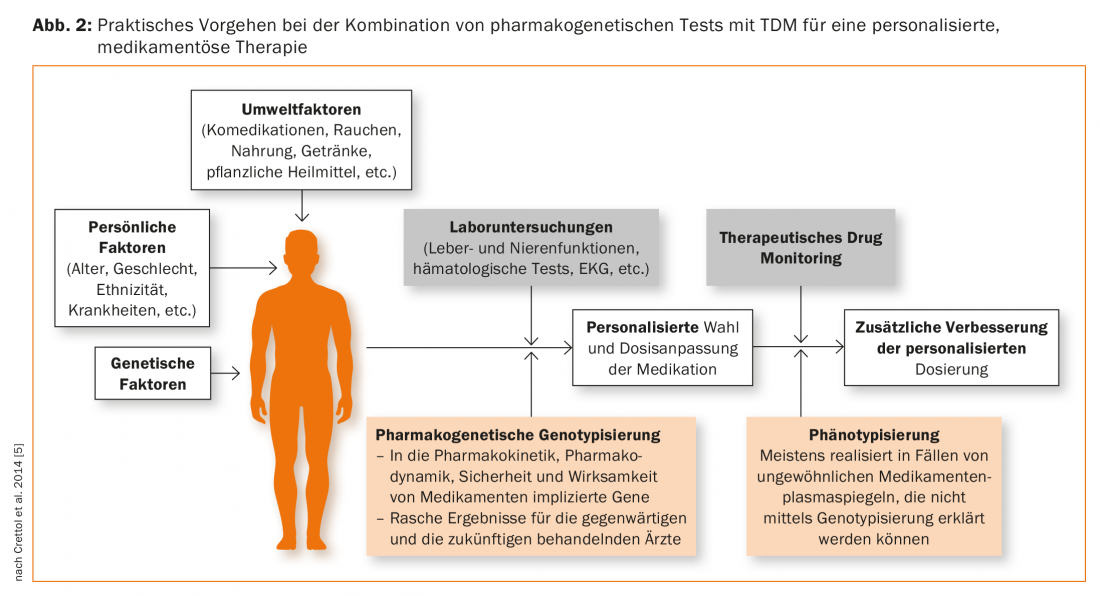
Genotyping leads to results that have lifelong validity and they therefore represent a kind of “trait marker”. The result of a phenotyping, such as the determination of a metabolic rate, on the other hand, is to be considered as a “state marker”. For this purpose, the patient is administered a substance which is the substrate of the enzyme to be tested, then blood or urine is collected depending on the protocol, then generally the parent substance and the metabolite are determined. The metabolite/mother substance ratio provides a metabolic quotient, which is a measure of the metabolizing capacity of the enzyme tested, as illustrated above using the example of the Debrisoquintest. The result can provide information on the patient’s genetic profile by EM and PM differing in metabolic quotient. However, if the patient is under the influence of drugs or other foreign substances that inhibit or induce the enzyme, this environmental influence masks the genetic component of the metabolism of the test substance. In psychiatry the following phenotyping is common: CYP2D6 (dextromethorphan as test substance), CYP2C19 (omeprazole), CYP1A2 (caffeine), CYP2B6 (bupropion), CYP3A (midazolam) [43,44]. Phenotyping is particularly useful for enzymes such as CYP1A2 and CYP3A (CYP3A4), which show large interindividual differences in activity but for which no clinically relevant genetic polymorphism is known and genotyping is therefore not useful. Fexofenadine [44] or digoxin [45] have been proposed for phenotyping ABCB1 activity. Depending on the question, several test samples for the different enzymes or transporters can be administered simultaneously as a “cocktail” [44,45]. Phenotyping is therefore complementary to TDM and genotyping (Fig. 2) [46], as the result provides information on the actual state of enzyme or transporter activity at the time of the assay [42].
Information such as drug plasma levels, comedications, comorbidities, nature of side effects if any, information on therapeutic response to medication, is helpful and necessary for the clinical pharmacologist to justify a prescription. For pharmacogenetic questions, the Swiss Centers for Clinical Pharmacology and Toxicology offer consultations, and patients can be referred for indication for pharmacogenetic testing on a consultative basis.
Conclusions
The time is now to move forward with the implementation of pharmacogenetic testing in psychopharmacotherapy. They can be a valuable tool to complement clinical investigations as well as TDM (Fig. 2) . Authorities such as the FDA and the EMA, as well as scientific bodies such as PharmGKB and CPIC, regularly publish recommendations for the clinical use of pharmacogenetic tests. Thus, there are now guidelines developed by independent bodies that are suitable for use in everyday clinical practice. They promote the proper implementation and interpretation of the results of pharmacogenetic tests and thus an optimized therapy depending on the genotype [48]. It is important that the genetic outcome is placed in the overall context of the patient’s therapy, i.e., that non-genetic influences such as comedication are considered when adjusting therapy. However, regionally adapted guidelines are now also necessary in order to enable an economically and clinically meaningful application adapted to local conditions following the political decision to make pharmacogenetic tests subject to certain conditions subject to compulsory health insurance and thus more accessible. Improper application may lead to disappointing results and the practicing physician will lose confidence in pharmacogenomics and personalized therapy in the medium term [49]. As mentioned above, the topic of “pharmacogenomics in psychiatry” should be increasingly considered in the education and training of specialists in psychiatry and psychotherapy [37–39]. A promotion of the cooperation between psychiatrists and practicing physicians on the one hand and the specialist in clinical pharmacology on the other hand is therefore to be striven for. At the same time, there are still many unresolved questions, which is why adequately planned and conducted clinical studies on pharmacogenetics in particular are still needed.
Literature:
- Alexanderson B, Evans DA, Sjöqvist F: Steady-state plasma levels of nortriptyline in twins: influence of genetic factors and drug therapy. British Medical Journal 1969; 4(686): 764-768.
- Mahgoub A, et al: Polymorphic hydroxylation of debrisoquine in man. Lancet 1977: 584-6.
- Gonzalez FJ, et al: Characterization of the common genetic defect in humans deficient in debrisoquine metabolism. Nature 1988; 331: 442-446.
- Bertilsson L, et al: Slow hydroxylation of nortriptyline and concomitant poor debrisoquine hydroxylation: clinical implications. Lancet 1981; i: 560-561.
- Crettol S, et al: Pharmacogenomics in psychiatry: from therapeutic drug monitoring to genomic medicine. Clin Pharmacol Ther 2014; 95(3): 254-257.
- Pratt V, et al: Medical Genetics Summaries. Bethesda (Md), USA: National Center for Biotechnology Information; 2017. 416 p.
- Zhang G, Nebert DW: Personalized medicine: genetic risk prediction of drug response. Pharmacol Ther 2017; 175: 75-90.
- Relling MV, Evans WE: Pharmacogenomics in the clinic. Nature 2015; 526(7573): 343-350.
- Patil J: Pharmacogenetics and Pharmacogenomics: A Brief Introduction. Journal of Pharmacovigilance 2015; 3: e139.
- Sim SC, Ingelman-Sundberg M: Pharmacogenomic biomarkers: new tools in current and future drug therapy. Trends Pharmacol Sci 2011; 32(2): 72-81.
- Dingermann T, Zündorf I: Stratified pharmacotherapy. Genetic principles, practical procedure. Eschborn, Germany: Govi; 2017. 339 p.
- Gandal MJ, et al: The road to precision psychiatry: translating genetics into disease mechanisms. Nat Neurosci 2016; 19(11): 1397-1407.
- Lohoff FW, Ferraro TN: Pharmacogenetic considerations in the treatment of psychiatric disorders. Expert Opin Pharmacother 2010; 11(3): 423-439.
- Budde M, et al: Pharmacogenomic aspects of bipolar disorder: An update. Eur Neuropsychopharmacol 2017; 27(6): 599-609.
- Malhotra AK, Zhang JP, Lencz T: Pharmacogenetics in psychiatry: translating research into clinical practice. Mol Psychiatry 2012; 17(8): 760-769.
- Wolking S, et al: Impact of Genetic Polymorphisms of ABCB1 (MDR1, P-Glycoprotein) on Drug Disposition and Potential Clinical Implications: Update of the Literature. Clinical Pharmacokinetics 2015; 54(7): 709-735.
- Caudle KE, et al: Standardizing terms for clinical pharmacogenetic test results: consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC). Genet Med 2017; 19(2): 215-223.
- Hicks JK, et al: Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin Pharmacol Ther 2017; 102(1): 37-44.
- Gasche Y, et al: Codeine intoxication associated with ultrarapid CYP2D6 metabolism. N Engl J Med 2004; 351(27): 2827-2831.
- McCormack M, et al: HLA-A*3101 and carbamazepine-induced hypersensitivity reactions in Europeans. N Engl J Med 2011; 364(12): 1134-1143.
- de Leon J: Translating Pharmacogenetics to Clinical Practice: Do Cytochrome P450 2D6 Ultrarapid Metabolizers Need Higher Atomoxetine Doses? J Am Acad Child Adolesc Psychiatry 2015; 54(7): 532-534.
- Whirl-Carrillo M, et al: Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther 2012; 92(4): 414-417.
- McDonagh EM, et al: From pharmacogenomic knowledge acquisition to clinical applications: the PharmGKB as a clinical pharmacogenomic biomarker resource. Biomark Med 2011; 5(6): 795-806.
- Dickmann LJ, Ware JA: Pharmacogenomics in the age of personalized medicine. Drug Discov Today Technol 2016; 21-22: 11-6.
- Stingl JC, Brockmoller J, Viviani R: Genetic variability of drug-metabolizing enzymes: the dual impact on psychiatric therapy and regulation of brain function. Mol Psychiatry 2013; 18(3): 273-287.
- Stingl JC, Brockmoller J: [Personalised pharmacogenetics. Evidence-based guidelines and clinical application of pharmacogenetic diagnostics]. Bundesgesundheitsblatt, Health Research, Health Protection. 2013; 56(11): 1509-1521.
- Ravyn D, et al: CYP450 pharmacogenetic treatment strategies for antipsychotics: a review of the evidence. Schizophr Res 2013; 149(1-3): 1-14.
- Fabbri C, et al: Progress and prospects in pharmacogenetics of antidepressant drugs. Expert Opin Drug Metab Toxicol 2016; 12(10): 1157-1168.
- Spina E, de Leon J: Clinical applications of CYP genotyping in psychiatry. J Neural Transm (Vienna) 2015; 122(1): 5-28.
- Holsboer-Trachsler E, et al: Traitement aigu des épisodes dépressifs. Swiss Medical Forum 2016; 16(35): 716-724.
- Uhr M, et al: Polymorphisms in the drug transporter gene ABCB1 predict antidepressant treatment response in depression. Neuron 2008; 57(2): 203-209.
- Breitenstein B, et al: ABCB1 gene variants and antidepressant treatment outcome: A meta-analysis. Am J Med Genet B Neuropsychiatr Genet 2015; 168B(4): 274-283.
- Breitenstein B, et al: Association of ABCB1 gene variants, plasma antidepressant concentration, and treatment response: results from a randomized clinical study. J Psychiatr Res 2016; 73: 86-95.
- Bruckl TM, Uhr M: ABCB1 genotyping in the treatment of depression. Pharmacogenomics 2016; 17(18): 2039-2069.
- Bschor T, et al:[Genetic tests for controlling treatment with antidepressants]. Neurologist 2017; 88(5): 495-499.
- O’Brien FE, et al: Human P-glycoprotein differentially affects antidepressant drug transport: relevance to blood-brain barrier permeability. Int J Neuropsychopharmacol 2013; 16(10): 2259-2272.
- Baumann P, et al: A proposal for a psychopharmacology-pharmacotherapy catalogue of learning objectives and a curriculum in Europe. World J Biol Psychiatry 2017; 18(1): 29-38.
- Whirl-Carrillo M, et al: Novel Disease-Drug Database Demonstrating Applicability for Pharmacogenomic-Based Prescribing. Clin Pharmacol Ther 2016; 100(6): 600-602.
- Collins SL, Carr DF, Pirmohamed M: Advances in the pharmacogenomics of adverse drug reactions. Drug Saf 2016; 39(1): 15-27.
- Green ED, Guyer MS; National Human Genome Research I: Charting a course for genomic medicine from base pairs to bedside. Nature 2011; 470(7333): 204-213.
- Hiemke C, et al: AGNP consensus guidelines for therapeutic drug monitoring in psychiatry: update 2011. Pharmacopsychiatry 2011; 44(6): 195-235.
- Hiemke C, et al: Consensus Guidelines for Therapeutic Drug Monitoring in Neuropsychopharmacology: Update 2017. Pharmacopsychiatry 2017 Sep 14. doi: 10.1055/s-0043-116492. [Epub ahead of print]
- Hiemke C, Shams M: Phenotyping and genotyping of drug metabolism to guide pharmacotherapy in psychiatry. Curr drug deliv 2013; 10(1): 46-53.
- Bosilkovska M, et al: Geneva cocktail for cytochrome p450 and P-glycoprotein activity assessment using dried blood spots. Clin Pharmacol Ther 2014; 96(3): 349-359.
- Fuhr U, Jetter A, Kirchheiner J: Appropriate phenotyping procedures for drug metabolizing enzymes and transporters in humans and their simultaneous use in the “cocktail” approach. Clin Pharmacol Ther 2007; 81(2): 270-283.
- Crettol S, et al: Pharmacogenomics in psychiatry – from TDM to genomic medicine. Clin Pharmacol Ther 2014; 95(3): 254-257.
- Baumann P, et al: Epileptiform seizure following sertraline treatment in an adolescent suffering from obsessive compulsive disorder and presenting a rare pharmacogenetic status. J Clin Psychopharmacol 2006; 26(6): 679-681.
- Amstutz U, Carleton BC: Pharmacogenetic testing: time for clinical practice guidelines. Clin Pharmacol Ther 2011; 89(6): 924-927.
- de Leon J: Pharmacogenetic Tests in Psychiatry: From Fear to Failure to Hype. J Clin Psychopharmacol 2016; 36(4): 299-304.
- Mlakar V, et al: Pharmacogenomics in Pediatric Oncology: Review of Gene-Drug Associations for Clinical Use. Int J Mol Sci 2016; 17(9): 1502.
InFo NEUROLOGY & PSYCHIATRY 2017; 15(6): 21-30.