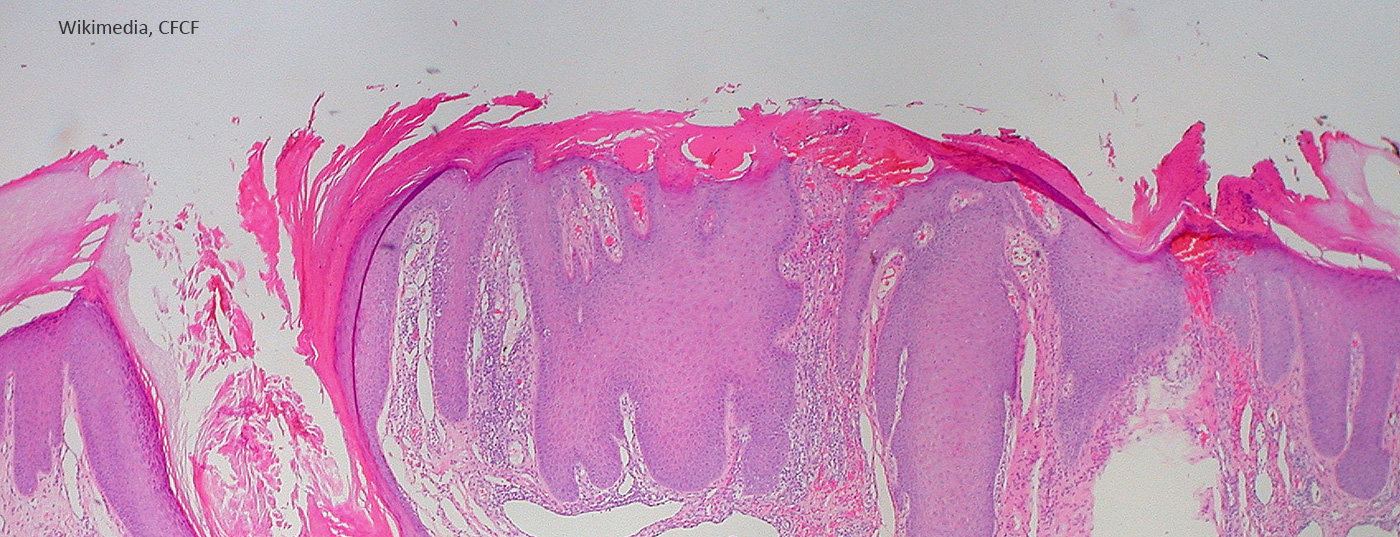
The question of which mechanisms contribute to the chronification of pruritus in chronic nodular prurigo is the subject of current research. It is hoped that new insights will be gained from the investigation of molecular processes. In a project presented at this year’s ADF meeting, DNA methylation patterns of pruriginous-lesional and non-lesional skin of prurigo patients were analyzed and compared to a skin-healthy control group.
Prurigo nodularis is defined as the presence of numerous, symmetrically distributed, hyperkeratotic or erosive nodules, especially in regions readily accessible for scratching. The predominant symptom is severe itching [1]. Persistent pruritus, sometimes accompanied by secondary scratch lesions, is referred to as chronic nodular prurigo. It is still unclear whether scratching per se is responsible for the development and maintenance of chronicity by inducing the itch-scratch cycle, or whether other mechanisms are also involved [2].
Analysis of epigenetic mechanisms
To find out more, a research team from Münster University Hospital analyzed the DNA methylation patterns of five patients with chronic nodular prurigo (CNPG) and compared them with a skin-healthy control group [2]. DNA methylation profiles can be used to identify differences between the DNA of diseased and healthy tissue. DNA methylation is a relatively stable modification of the epigenome.
One aim of the present study was to find biomarkers based on DNA methylation that would allow conclusions to be drawn about the mechanisms underlying the chronicity of pruritus. For this purpose, biopsies of pruriginous lesional (PL) and non-pruriginous non-lesional (NPNL) skin from patients and healthy controls were analyzed using Illumina Infinium Methylation EPIC arrays (Diagonede, Belgium). To identify similar subgroups in the dataset based on differential DNA methylation, distance clustering analysis was then performed, followed by principal component analysis (PCA).

Differences in DNA methylation could be detected
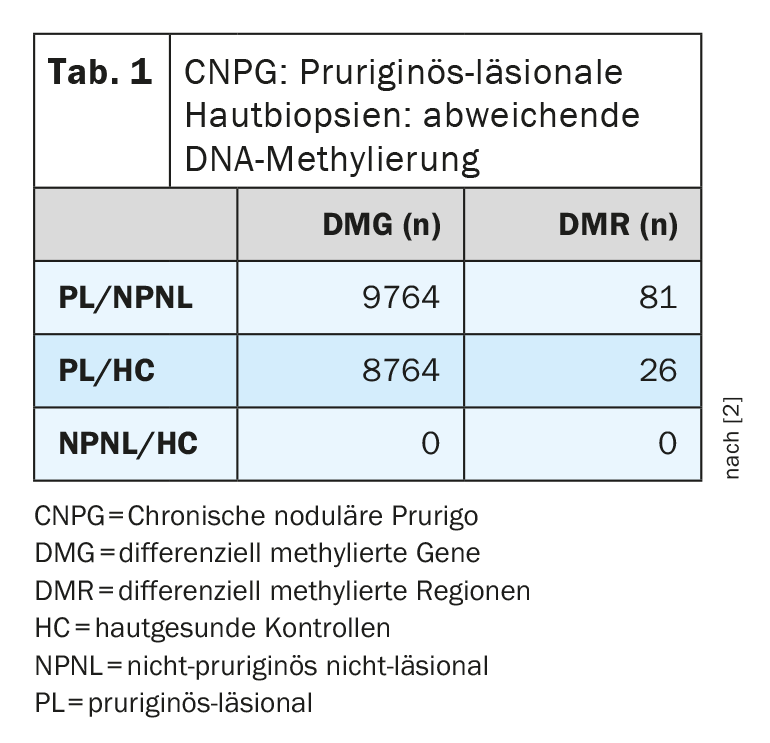
The results show that both distance clustering and PCA were able to differentiate pruriginous-lesional (PL) from non-pruriginous-non-lesional (NPNL) biopsies and from skin-healthy controls (HC) [2]. The researchers found a significant number of differentially methylated positions (DMPs) between PL and NPNL (n=28879) and between PL and HC (n=24994). This highlights the DNA methylation differences that the researchers suspect. In contrast, no differentially methylated positions (DMPs) or regions (DMRs) could be detected when comparing NPNL and HC ( Table 1) .
In summary, the present study identified characteristic clusters of DNA methylation depending on lesion status in CNPG. Further analyses regarding correlations with gene expression profiles are currently underway and may contribute to a deeper understanding of the mechanisms leading to chronicity of pruritus.
Congress: Working Group of Dermatological Research
Literature:
- Mettang T, Vonend A, Raap U: Prurigo nodularis in dermatoses and systemic diseases. Dermatologist 2014; 65(8): 697-703.
- Agelopoulos K, et al: Pruritic lesional skin in chronic nodular prurigo exhibits specific DNA methylation signatures, P188, ADF Annual Meeting, 23-26 Feb 2022.
- Manjrekar J. Epigenetic inheritance, prions and evolution. J Genet 2017; 96(3): 445-456.
- “Successful research on chronic pruritus continues: DFG research group enters second funding phase”, Münster University Hospital, 30.09.2021
- Wieser JK, Mercurio MG, Somers K: Resolution of Treatment-Refractory Prurigo Nodularis With Dupilumab: A Case Series Cureus 2020; 12(6): e8737.
DERMATOLOGIE PRAXIS 2022; 32(2): 24 (published 4/20/22, ahead of print).