How can kinase inhibitors be put to good use? What are the advantages or disadvantages of a combination? How can genomic sequencing contribute to an individualized therapeutic approach with such inhibitors and which study designs should be used to test this? What can be learned from failed studies and development processes of known kinase inhibitors? All these questions were discussed at the ESMO Congress in Amsterdam.
Annette K. Larsen, Paris, asked whether and how to combine kinase inhibitors:
“The development of new anticancer agents that target oncogenic signaling pathways represents a major conceptual breakthrough. Nevertheless, clinical results have often fallen short of expectations, in part due to downstream mutations, unexpected feedback loops, or so-called cross-talk of signaling pathways.”
For this reason, there is a strong focus today on targeting either multiple signal paths simultaneously or different steps in the same signal path. Often, kinase inhibitors were added directly to established cytotoxic agents without dose adjustment, sometimes resulting in severe toxic side effects. In her presentation, Larsen outlined the possibilities and limitations of kinase inhibitor combinations.
Two recent phase III trials (PACCE and CAIRO2) evaluated the addition of EGFR-targeted monoclonal antibodies (mAbs), cetuximab and panitumumab, respectively, to bevacizumab plus chemotherapy in patients with colorectal cancer (CRC). This combination was associated not only with shorter progression-free survival but also with poorer quality of life, again in patients with wild-type KRAS tumor.
“Why did the combination of the two mAbs not work? First, the combination of target agents was too toxic, and second, it is not active, meaning both inhibit extracellular ligands or receptors but have limited, perhaps no, effects on receptor tyrosine kinase (RTK) signaling,” Larsen said. It concludes that although the combination of VEGF- and EGFR-targeted agents is possible, it does not necessarily have to be used at the same time as chemotherapy. KRAS mutation status plays a role here, especially in EGFR-targeted mAbs.
Individualized therapy approaches
Prof. Emile E. Voest, M.D., Utrecht, spoke about the possibilities of including genomic sequencing in therapeutic decision-making: “Detailed information on how a tumor is genetically derived enables more targeted selection of specific patients for a given therapy. Examples such as trastuzumab for HER2 expressions in breast cancer, imatinib for BCR-ABL translocations in leukemia, vemurafenib and crizotinib against V600E mutations in melanoma, and ALK-EML4 translocations in lung cancer, respectively, have clearly demonstrated how validly the concept can be applied. These spectacular successes are clouded by the fact that they are temporary because resistance often develops.”
Thanks to advances in sequencing technology, it is now possible to generate detailed information about the genomic abnormalities in a tumor. Single-gene analyses (i.e. BRAF, KRAS) will be replaced by whole-genome analyses in the near future. This global approach allows insights at the pathway level rather than at the individual gene level. “For example, in breast cancer patients, we could not find a correlation between the success of chemotherapy and specifically trackable mutations in the PI3K gene, but we could very much find one to mutations in the PI3K pathway,” Prof. Voest said.
Tumor heterogeneity is an important dimension of tumor growth and clonal outgrowth of resistant populations often occurs during treatment. Here, “(ultra)deep sequencing” methods can help to detect and identify the clones early, allowing an anticipatory treatment approach. Current thinking is that it will be easier to find a predictive genetic profile for drugs with a specific mechanism of action (such as vemurafenib) than for broad-spectrum kinase inhibitors (such as sunitinib). Considering the findings on the above-mentioned correlation between the success of chemotherapy and the PI3K pathway, this approach may be exposed as a misunderstanding.
“We are at the beginning of an era in which comprehensive genetic testing of the tumor and germline DNA will be part of regular diagnostics in cancer patients. In particular, the competent interpretation of the extremely complex data will be crucial for the correct DNA-guided therapy selection. This will require large databases that offer the possibility of linking clinical success with genetic data,” Prof. Voest concluded his presentation.
Genotype vs. basket studies
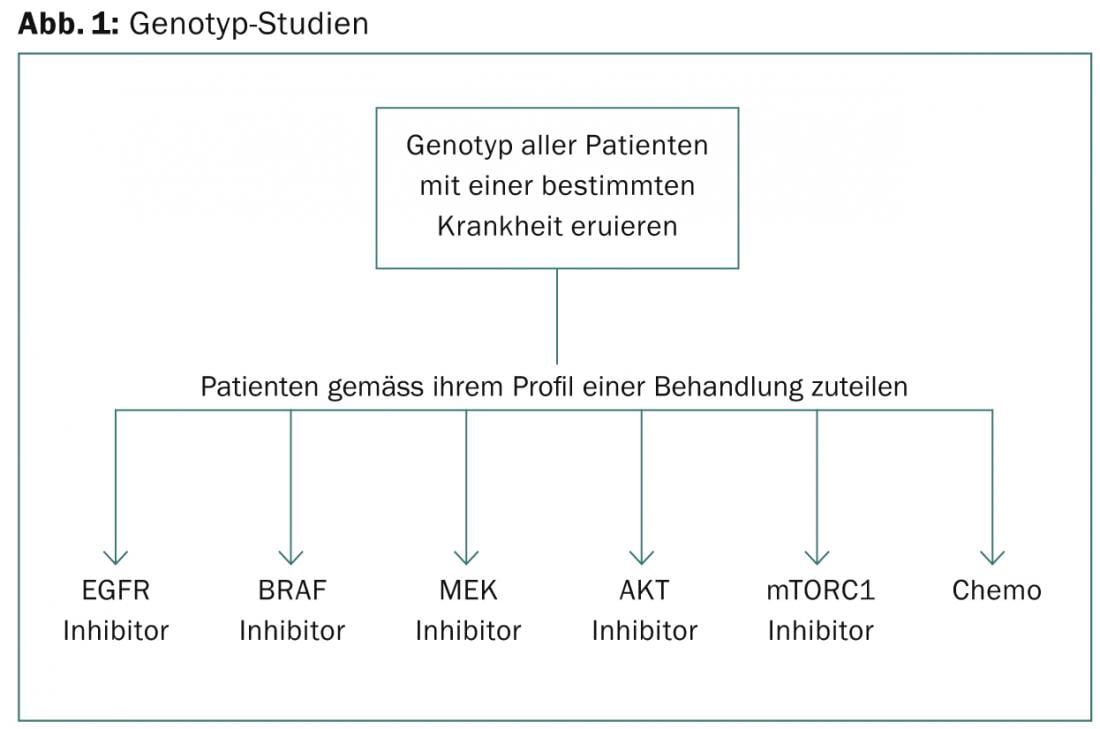
“Genotype studies are based on screening a certain number of patients with a disease for their genotype and then assigning them to different targeted drugs according to their profile,” said José Baselga, MD, New York ( Fig. 1). “The problems with this approach are as follows: The drugs used in each group are usually not best-in-class, but available (e.g., the first BATTLE trial used sorafenib as the RAF inhibitor). Moreover, if randomization is used, it might seem unethical during the course of the trial to randomize some patients who are thus not treated according to their profile. Moreover, the total number of participants is usually low, therefore this design can often acquire too few patients with rare mutations to validly compare clinical outcome with genetic profile (e.g. BRAF in lung cancer or ERBB2 mutations in ovarian cancer).”
So-called. Basket studies, in contrast, allow testing of a specific hypothesis, e.g.: Do patients with biliary cancer whose tumor harbors BRAF(V600E) mutations respond to vemurafenib? Furthermore, because this type of study is based on a collection of disease-specific cohorts, it can be used to examine the influence of ancestry on drug response. Furthermore, tissue sampling may explain response heterogeneity. The primary criticism of basket studies is that they may miss those patients who might respond but do not have the biomarker they are looking for. In addition, identification of participants remains a hurdle: According to Baselga, it is therefore imperative to separate the screening protocol from the treatment protocol.
Learning from mistakes
How can we learn from failed kinase inhibitor trials? This question was posed by Stefan Sleijfer, MD, Rotterdam: “In recent years, several new kinase inhibitors have been rapidly and successfully approved on the basis of sometimes sensational study results. In contrast, however, many such agents also failed, sometimes at an early clinical stage, sometimes only after extensive and expensive phase III trials had been conducted. Valuable conclusions for future study designs can be drawn from both the successful and failed development processes.”
The most successful kinase inhibitors are those that directly target and inhibit the product of a mutated gene, thus affecting a specific subset of a disease. Once the mechanism of action of these drugs was known and thus eligible patients could be specifically selected, the drugs were clinically tested in specifically selected patient groups from the outset. Successful examples include imatinib in malignant gastrointestinal stromal tumors (GIST), vemurafenib in BRAF(V600E) mutant melanoma, or crizotinib in non-small cell lung cancer (NSCLC) with EML4-ALK fusion gene.
An important common characteristic of those drugs that failed or achieved approval only after an arduous and expensive path is that the mechanism of action was unknown before the start of clinical trials. Therefore, predictive profiles could not be established, patients likely to respond could not be identified a priori, and clinical trials were conducted in unselected patient populations. Current examples include IGF-1R antagonists and mTOR inhibitors.
“The most important lesson of these studies, then, is the utility and need to uncover in preclinical settings the exact mechanism of action of the drug under investigation and to identify predictive markers before large clinical trials are initiated. If the mechanism is unknown, it remains questionable whether the particular drug should enter the development process. Especially because this requires an extremely large amount of resources and money that could be better spent on other compounds to be tested. If a drug with an unknown mechanism of action is nevertheless already in development, adaptive designs can be used. For example, biomaterial collection is essential to retrospectively uncover predictive profiles, as was the case with EGFR kinase inhibitors in NSCLC,” Dr. Sleijfer concluded.
Source: “Optimal use of Targeted Kinase Inhibitors (TKIs),” Symposium at ESMO Congress, September 27 – October 1, 2013, Amsterdam.
InFo ONCOLOGY & HEMATOLOGY 2014; 2(1): 45-47.